Research
Our team is developing new methods for single-molecule DNA sequencing. We are interested in how genes work and would like to study the way in which proteins change DNA structure to switch genes on and off. We are also pursuing the chemistry and physics of the liquid-solid interface and are trying to further the development and improvement of nanobiophysical techniques (in particular the combination of cutting-edge force and optical technologies) and their application to fundamental biological processes related to mechanical forces such as cell adhesion, cell and tissue mechanics and cell interactions, as well as biomolecular interactions and conformations.
Capabilities
Areas of investigation include:
Cell adhesion
Single cell force spectroscopy is a method to quantitatively study the interaction of a single cell with surfaces or other cells. For these experiments, a single cell is attached to the cantilever of an atomic force microscope, and the interaction forces between this cell and surfaces or other cells are probed with piconewton precision.
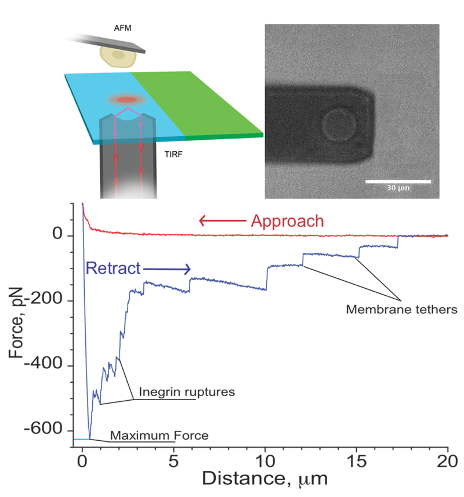
I. Yermolenko, A. Fuhrmann, S. Magonov, V. Lishko, S. Oshkadyerov, R. Ros, and T. Ugarova; Origin of the Nonadhesive Properties of Fibrinogen Matrices Probed by Force Spectroscopy, Langmuir 26 17269–17277 (2010).
I.S. Yermolenko, O.V. Gorkun, A. Fuhrmann, V.K. Lishko, S.P. Oshkadyerov, S.T. Lord, R. Ros, and T.P. Ugarova; The assembly of nonadhesive fibrinogen matrices depends on the αC regions of the fibrinogen molecule, J. Biol. Chem. 287 41979-41990 (2012).
W. Christenson, I. Yermolenko, B. Plochberger, F. Camacho-Alanis, A. Ros, T.P. Ugarova, and R. Ros; Combined single cell AFM manipulation and TIRFM for probing the molecular stability of multilayer fibrinogen matrices, Ultramicroscopy 136 211–215 (2014).
R. Safiullin, W. Christenson, H. Owaynat, I.S. Yermolenko, M.K. Kadirov, R. Ros and T. P. Ugarova; Fibrinogen matrix deposited on the surface of biomaterials acts as a natural anti-adhesive coating Biomaterials67, 151-159 (2015).
Cell and tissue mechanics
Cell behavior is guided by the 3D microenvironment. Reciprocal mechanical interactions between cells and their microenvironment can dictate cell phenotype and behavior, requiring studies of cells in physiologically relevant 3D extracellular matrices (ECM). In cancer development, cell and matrix stiffness has been demonstrated to be a key indicator of metastatic potential. To assess the mechanical interplay between the cells and ECM during invasion, we combined confocal fluorescence microscopy and atomic force microscopy (AFM) indentation to determine the viscoelastic properties of individual embedded cells and the pericellular matrix using novel analysis methods for heterogeneous samples.
A. Fuhrmann, J. R. Staunton, V. Nandakumar, N. Banyai, P. Davies, and R. Ros; AFM stiffness nanotomography of normal, metaplastic and dysplastic human esophageal cells, Physical Biology 8 015007 (2011).
J. Staunton, B. Doss, S. Lindsay, R. Ros; Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen I matrices Scientific Reports 6:19686 (2016).
N. Peela, F.S. Sam; W. Christenson, D. Truong, A.W. Watson, G. Mouneimne, R. Ros, and M. Nikkhah; A Three Dimensional Micropatterned Tumor Model for Breast Cancer Cell Migration Studies, Biomaterials 81, 72-83 (2016).
H. Saini, K. Rahmani Eliato, C. Silva, M. Allam, R. Ros, M. Nikkhah; The Role of Desmoplasia and Stromal Fibroblasts on Anti-Cancer Drug Resistance in a Microengineered Tumor Model, Cellular and Molecular Bioengineering, 11, 5, 419–433 (2018)
B.L. Doss, K.Rahmani Eliato, K.-h. Lin, and R. Ros; Quantitative mechanical analysis of indentations on layered, soft elastic materials, Soft Matter 15, 1776-1784 (2019).
Saini, K. Rahmani Eliato, J. Veldhuizen, A. Zareb, M. Allam, C. Silva, A. Kratz, D. Truong, G. Mouneimne, J. LaBaer, R. Ros, M. Nikkhah; The role of tumor-stroma interactions on desmoplasia and tumorigenicity within a microengineered 3D platform, Biomaterials 247, 119975 (2020).
A. Buchberger, H. Saini, K. Rahmani Eliato, R. Merkley, Y. Xu, A. Zare, J. Bernal, M. Nikkhah, R. Ros, N. Stephanopoulos; Reversible Control of Gelatin Hydrogel Stiffness using DNA Crosslinkers, ChemRxiv. Preprint https://doi.org/10.26434/chemrxiv.12461918.v1 (2020).
Electronic sequencing of biomolecules
DNA testing is transforming health care and medicine, but current technologies only give a snapshot of an individual’s genetic makeup. Current sequencing techniques are relatively slow and expensive.
Our approach is making direct electrical measurements on the DNA itself, or on the molecule that copies DNA, DNA polymerase. The hope is that a small computer-chip sized device could sequence a human genome in hours.
Direct electrical measurements of protein motion enable a range of applications. For example, CRISPR associated proteins can be followed in real time as they search for, and then find their single molecule targets. Other important molecules, such as biological sugars and proteins themselves can be analyzed with this new technology. This work is funded by the National Institute for Human Genome Research and carried out in collaboration with Xu Wang of the ASU School of Molecular Sciences.
Electron transfer of single molecules
We are using both the scanning tunnel microscope and a novel STM-atomic force microscope combination instrument to probe electron transfer in single molecules. This is important for understanding processes such as photosynthesis and for building molecular electronic devices.
To do this, we need to be able to make a variety of interesting molecules and tether them to a conducting substrate. We are working with Tom and Anna Moore and Devens Gust of the ASU Center for Early Events in Photosynthesis.
We are also studying proteins as molecular wires and electric components in collaboration with Giovanna Ghirlanda in the School of Molecular Sciences.
The tools behind the technology development are:
- Engineering and chemistry: nanostructures that connect to molecules and instruments that manipulate and measure at the single molecule level.
- Physics: understanding the quantum phenomena that dominate interactions on this length scale and building simple models of the complex many-particle phenomena.
- Biology: molecules involved in gene expression (signaling and the control of processes in living cells).
- Finally, the building of molecular model systems and the controlled assembly of nanostructures is dependent upon chemistry, biochemistry and materials chemistry.
Nonlinear super-resolution microscopy
Progress in microscopy techniques has been a key driver for new findings in biomedical sciences. Most prominently, fluorescence microscopy combines the highest sensitivity with molecular specificity and excellent image contrast. Over the last decades, the spatial resolution limit set by the diffraction of light (typically ~200 nm laterally and ~500 nm axially at a wavelength of 500 nm) has been overcome by the development and evolution of new so-called super‐resolution fluorescence microscopy methods. However, until now non-linear microscopy techniques hardly profited from these super-resolution methods. Among the non-linear microscopy techniques are two-photon excitation (2PE) microscopy, second harmonic generation (SHG) imaging, and third harmonic generation (THG) imaging. We recently demonstrated the proof-of-concept for a 2PE and SHG super-resolution technique based on Image Scanning Microscopy (ISM).
I. Gregor, M. Spiecker, R. Petrovsky, J. Großhans, R. Ros, and J. Enderlein; Rapid Non‐linear Image Scanning Microscopy, Nature Methods 14, 1087–1089 (2017).
Research in chromatin and biomolecular structure
We would like to understand how genes work at a molecular level. Specifically, what is it about the binding of regulatory proteins (and other chemicals) that switches certain genes on and others off? It is here that the secrets of development (i.e., what makes one cell a red-blood cell and another a neuron?) and the lifecycle of the cell lie.
The scanning probe microscopes, capable of imaging in fluid at very high resolution, offer the chance of imaging the components of a regulatory complex (DNA and protein) in natural conditions and possibly even in action (that is, it might be possible to make movies of biological processes at the molecular level).
We have developed more sensitive instruments capable of gentler imaging at higher resolution, and we have used them to study structural transitions in DNA.
With this project, we aim to:
- Develop rapid and reliable antibody-based molecular recognition mapping.
- Characterize and test molecular recognition mapping on known and defined model chromatin.
- Carry out linear organization, higher order structures and molecular recognition mapping studies of long terminal repeat promoter region chromatinx.
- Study glucocorticoid receptor binding to this chromatin.
- Study chromatin remodeling in relation to higher order structures and molecular recognition mapping.
This work has been funded by the National Cancer Institute and is carried out in collaboration with Robert Ros who is affiliated with our center and the ASU Department of Physics.
Single molecule interactions
Binding forces between single receptor and ligand molecules can be investigated in dynamic force spectroscopy experiments by atomic force microscopy (AFM) or optical laser tweezers (OT) revealing details of the interaction mechanism, the kinetics of the reaction and the energy landscape of the binding. The ability to measure inter- and intramolecular forces and elasticities with pico-Newton-, nanometer- and millisecond resolution allows investigation of single ligand-receptor interaction in a broad affinity range for dissociation constants (KD) from 10-5 M to 10-15 M at a discrimination level of single point mutations. We applied this technique to a wide range of interactions including antibody-antigen, protein-DNA, protein-RNA, synthetic bioorganic interactions, supramolecular guest-host interactions, and single DNA base pairs.
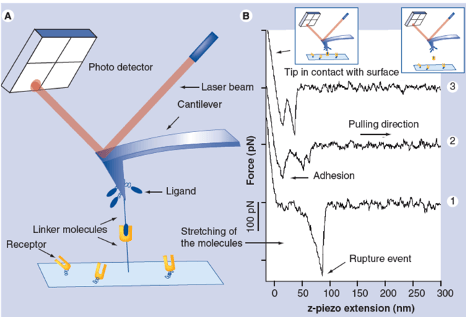
R. Ros, F. Schwesinger, D. Anselmetti, M. Kubon, R. Schäfer, A. Plückthun, L. Tiefenauer; Antigen binding forces of individually addressed single-chain Fv antibody molecules: Proc. Natl. Acad. Sci. USA 95, 7402-7405 (1998).
B. Baumgarth, F. W. Bartels, D. Anselmetti, A. Becker, and R. Ros; Detailed studies of the binding mechanism of the Sinorhizobium meliloti transcriptional activator ExpG to DNA: Microbiology 151, 259-268 (2005).
A. Fuhrmann, S. Getfert, D. Anselmetti, P. Reimann, and R. Ros; Refined procedure of evaluating experimental single-molecule force spectroscopy data, Phys. Rev. E 77, 031912 (2008).
A. Fuhrmann, J.C. Schoening, D. Anselmetti, D. Staiger, and R. Ros; Quantitative analysis of single molecule RNA-protein interaction Biophysical Journal 96, 5030-5039 (2009).
Fuhrmann, S. Getfert, Q. Fu, P. Reimann, S. Lindsay, and R. Ros; Long lifetime of hydrogen-bonded DNA basepairs by force spectroscopy, Biophysical Journal 102 2381-2390 (2012).
S. Senapati, S. Biswas, S. Manna, R. Ros, S. Lindsay, P. Zhang; A Y–Shaped Three-Arm Structure for Probing Bivalent Interactions of Protein Receptor-Ligand with AFM and SPR, Langmuir , 34, 23, 6930-6940 (2018).
Single molecule interactions by fluorescence
We investigate protein-protein and protein-DNA interactions using single-molecule fluorescence techniques, including sm-FRET, and fluorescence correlation spectroscopy. We have developed a methodology to characterize complex protein assembly pathways by studying fluorescence fluctuations of proteins in solution. We are interested in characterizing the effect of salts and small molecules on protein stability and oligomerization equilibrium. A current focus of our research is understanding the molecular details of how common cellular osmolytes modulate the oligomerization state of proteins.
“Potassium Glutamate and Glycine Betaine Induce Self-Assembly of the PCNA and b-Sliding Clamps”. Anirban Purohit, Lauren G. Douma, Linda B. Bloom, M. Levitus. Biophys. J. 2020.
“Electrostatic Interactions at the Dimer Interface Stabilize the E. coli beta Sliding Clamp “. Anirban Purohit, Jennifer K. England, Lauren G. Douma, Farzaneh Tondnevis, Linda B. Bloom, M. Levitus. Biophys. J. 2017. 113 794-804.
A. Fuhrmann, S. Getfert, D. Anselmetti, P. Reimann, and R. Ros; Refined procedure of evaluating experimental single-molecule force spectroscopy data, Phys. Rev. E 77, 031912 (2008).
“Intrinsic stability and oligomerization dynamics of DNA processivity clamps”. Binder, J. K., Douma, L.G., Ranjit, S., Kanno D.M., Chakraborty, M., Bloom, L.B., and Levitus M. Nucl. Acids. Res. 2014. 42, 6476-6486.
“Protein Oligomerization Monitored by Fluorescence Fluctuation Spectroscopy: Self-Assembly of Rubisco Activase”. Chakraborty,M., Kuriata, A.M., Henderson, J.N., Salvucci, M.E., Wachter, R.M.and Levitus M. Biophys. J. 2012. 103, 949-958.
Additional capabilities
- Single molecule electronic measurements
- Single molecule optical measurements
- High-resolution optical imaging of molecules
- Scanning probe microscopy of processes at the molecular level
- Correlated scanning probe/optical microscopy
- Nanorheology
- Single-cell force spectroscopy
Support the Biodesign Center for Single Molecule Biophysics.
Advancing single-molecule-scale research and translating discoveries into new applications for health and combating disease.