Surface Plasmon Resonance Imaging
Team
Mitch Magee, PhD, Ji Qiu, PhD, Lushen Song, PhD, Brianne Petritis, and Ian Shoemaker
Collaborator
Manuel Fuentes, PhD (Universidad de Salamanca, Spain)
In the post-genome era having sequenced the human genome, one of the most important pursuits is to understand the function of proteins. The study of biochemical activity of gene products has led to a better understanding of many cellular signaling pathways and their regulation. These pathways have served as a foundation to develop models for the pathogenesis of various diseases, including cancer. This in turn has led to the development of various diagnostics and improved therapeutics. In order to increase the speed with which we explore the molecular pathways of disease, we have embraced various genomics and proteomics approaches to study biology at large scale. Thus far, this effort has generated immense collections of data which enhanced our biochemical understanding of protein function. However, most current proteome-scale technologies are fairly crude end point assays that provide little quantitative information compared with standard biochemical approaches.
Protein signaling via protein interactions is central to virtually all cellular processes and diseases. Current methods to detect and characterize protein-protein interactions generally fall into two categories: low throughput, which obtain detailed information on protein interactions but operate on only a few proteins at once (e.g., surface plasmon resonance; SPR), or which are high throughput, which have high false positive and negative rates, and provide little to no quantitative information pertaining to the strength and rate of those interactions (e.g., yeast two hybrid).
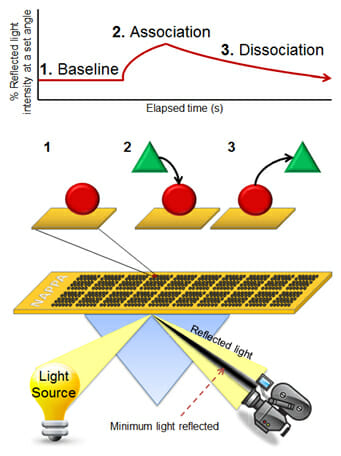
To address this issue, our lab has developed a high throughput approach that quantitatively characterizes more than 400 protein interactions simultaneously, collecting information on both interaction affinity and kinetics. In this platform, we have coupled our unique NAPPA protein microarray platform with SPR, enabling the assessment of affinities ranging from low micromolarity to high picomolarity. The SPRi method measures changes in refractive index very close to a sensor surface which are dependent on the dielectric properties of the metal, which in turn depend on what proteins are present on the surface (see image at left). This method allows for real time and label-free detection of binding events. Coupled with NAPPA, the format has been used to characterize networks of interactions in a signaling pathway, including affinities and kinetics. This open format allows for testing the effects of post-translational modifications (PTMs) by testing interactions in their presence and absence. It can also be used to explore the quantitative effects of numerous mutations and isoforms on the ability of a protein to interact with its targets. Two main projects that currently use this technology are the Physical Sciences in Oncology Center (PSOC) project to determine the kinetics of protein-protein interactions in the B-cell signaling pathway in Burkitt’s Lymphoma and in the NuProMer affinity reagent project to screen interactions between peptides and their target.