Technology
Nucleic Acid Programmable Protein Array
The NAPPA Protein Array Core uses a novel protein microarray technology, which replaces the complex process of spotting purified proteins with the simple process of spotting plasmid DNA. Our facility offers high throughput DNA preparation, pre-set and custom protein array production and array screening and analysis services. For more information, contact the core manager.
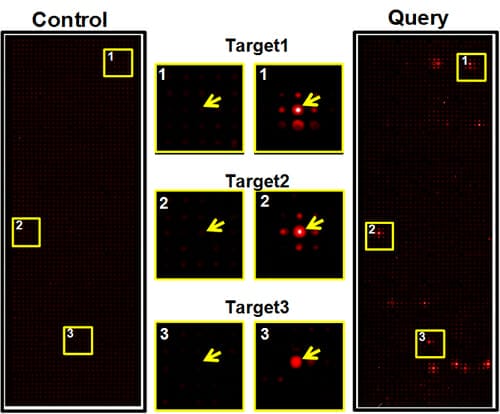
- Epitope mapping. Our Antigen-Specific Epitope-Targeting arrays will quickly and easily identify epitopes and enable rapid mapping antibody epitopes at the amino-acid level. Our approach will bypass the expensive cost of synthetic peptides, challenges of antigen purification, and the uncertainty of random peptide library screening. A nested series of deletion mutants, N-terminal, C-terminal, internal and tiling peptides can be generated rapidly and screened on ASET arrays at a low cost. Hundreds of arrays can be produced in one preparation and used for screening for many different antibodies to the target antigens. Our ASET arrays will allow epitope mapping in physiological buffers or under denaturing conditions.
- High-density arrays. Our high-density array development takes advantage of the mature semiconductor etching technology to produce silicon substrates with tens of thousands of nano-volume wells. Companion developments include a piezo-dispensing arrayer that allows the precise and accurate dispensation of genes into individual nano-wells, a filling device that allows the dispatch of nanoliter volumes of protein expression lysate into each well, and a sealing method that completely seals each micro-well to eliminate any well-to-well cross talk.
- High-sensitivity arrays. We have also been working with researchers at the company Meso Scale Discovery to reconfigure NAPPA to improve the sensitivity of signal detection. Meso Scale has created a new system that can detect signals using electrochemiluminescence, which means that the signal isn’t produced until stimulated by an electrochemical reaction. The benefit of this system is that it decreases background noise, and thus increases the signal to noise ratio, making it easier to detect binding of autoantibodies and differences between cases and control in NAPPA, for example. We are currently testing the NAPPA technology (i.e., spotting DNA and making protein in the Meso Scale wells) by spotting 4 spots of different DNA and detecting signals using electrochemiluminescence to see if the technologies are compatible. Future experiments will test if this system can be expanded for high throughput assays. If so, this system could significantly increase the sensitivity of our NAPPA assays to better identify biomarkers for disease and provide a potential tool for in the clinic assays.
- Surface Plasmon Resonance Imaging. Our lab has developed a high throughput approach that quantitatively characterizes more than 400 protein interactions simultaneously, collecting information on both interaction affinity and kinetics. In this platform, we have coupled our unique NAPPA protein microarray platform with SPR, enabling the assessment of affinities ranging from low micromolarity to high picomolarity. The SPRi method measures changes in refractive index very close to a sensor surface which are dependent on the dielectric properties of the metal, which in turn depend on what proteins are present on the surface. This method allows for real time and label-free detection of binding events. Coupled with NAPPA, the format has been used to characterize networks of interactions in a signaling pathway, including affinities and kinetics. This open format allows for testing the effects of post-translational modifications (PTMs) by testing interactions in their presence and absence. It can also be used to explore the quantitative effects of numerous mutations and isoforms on the ability of a protein to interact with its targets.
- Protein interactions. We have developed a high-throughput unbiased protein-protein interaction screening platform on NAPPA microarrays displaying 10,000 human proteins and an independent validation platform on magnetic beads using fluorescein labeled detection reagents with high-affinity and high-specificity. With these tools, we are performing a series of proteomics studies to characterize bacterial pathogen/virus and host interactions and identify the novel host targets which may play important roles in cancers and infectious diseases.
Post-translational modifications. NAPPA can be coupled with a variety of biochemical modification systems to identify proteins that add post-translational modifications to other proteins or to identify protein targets. Furthermore, once developed, these systems provide an ideal method for screening drugs or other small molecule inhibitors. We have three main projects studying different post-translational modifications:
o Phosphorylation and its applications in chronic myelogenous leukemia
o AMPylation and its role in bacterial pathogenesis
o Citrullination and its role in the pathogenesis of rheumatoid arthritis
Automation
Our team automates processes in the study of proteomics to speed up discovery. Important tools for this include our DNA factory and high-throughput cloning.
- DNA factory. The DNA factory was developed in collaboration with HighRes Biosolutions. This unique system allows us to prep 4,600 plasmids in 72 hours with limited technician intervention. The system contains three fully articulated robotic arms, eight 37°C shaking incubators, one dual arm liquid handler, two automated centrifuges, two automated plate sealers, two bulk reagent dispensers, one automated plate peeler, one -20°C incubator, one ambient plate storage hotel, one six mode plate washer, and one plate reader. All devices are controlled by scheduling software and data is real-time tracked to internal and off-line database laboratory information management systems.
- High-throughput cloning. Our High Throughput Cloning/Production Team works on full-length sequence validation of over 150,000 Protein Structure Initiative clones from NIGMS-funded PSI centers. In addition, our team does all of the recombinational cloning and clone validation projects both within the center and with numerous collaborators. Each step of our cloning, sequencing and distribution process is tracked by laboratory information management systems and facilitated by robots. All plasmids validated and created by the cloning team can be found in the DNASU Plasmid Repository. For more information and to order, visit DNASU.
Informatics
Our automation relies on advanced informatics, including sequence analysis, sample and NAPPA tracking, advanced literature mining and next-generation sequencing.
- Sequence analysis – ACE. Designed to manage thousands of clones simultaneously, ACE uses an Oracle database to store information about clones at various completion stages, project processing parameters, and acceptance criteria. The software has been used successfully to evaluate over 250,000 clones at the LaBaer lab. Its use has allowed a dramatic reduction in the amount of time and labor required to evaluate clone sequences and has decreased the number of missed sequence discrepancies, which commonly occur during manual evaluation. In addition, ACE has helped to reduce the number of sequencing reads needed to achieve adequate coverage for making decisions on clones.
- Sample tracking – FLEX. We have developed a laboratory information management system (LIMS) called FLEXGeneDB (FLEX) for tracking high throughput cloning processing, which is a major production pipeline in the lab. The FLEX system tracks every step in the production process and integrates with liquid handling robots to eliminate human errors. It maintains a detailed history for each plate and sample by capturing processing parameters, protocols and analytical results for the complete life cycle of the sample. FLEX is capable of managing clones produced outside our lab that require further processing such as transferring to different vectors or sequence validation. Visit the FLEX website.
- Advanced literature mining. High-throughput technologies, such as proteomic screening and DNA micro-arrays, produce vast amounts of data requiring comprehensive analytical methods to decipher the biologically relevant results. One approach would be to manually search the biomedical literature; however, this would be an arduous task. We developed two automated literature-mining tools, termed MedGene and BioGene, which can be used to comprehensively search the literature for associations between human genes and disease or biological themes, respectively.
- Next-generating sequencing. Our next generation sequencing service is now overseen by the ASU Genomics Facility.
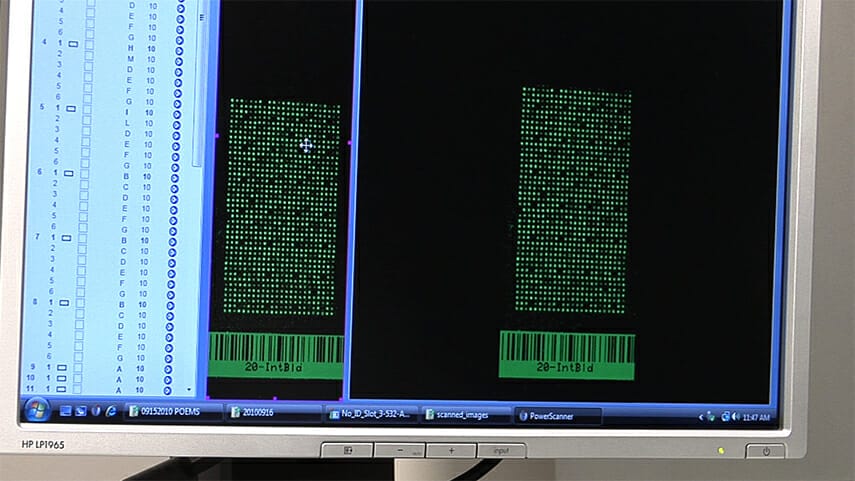
- NAPPA tracking. The web-based NAPPA Tracking System manages key data and information throughout the NAPPA process including: sample management, slide production, experiment results, data analysis and visualization. The sample management module provides direct access to the DNASU Repository and FLEXgene database to access plasmids targeted for the NAPPA microarray platform. Processes to grow the clones on 96-well plates, store glycerol stocks on 96-well plates (for re-growth later), prepare and quantify DNA are tracked and detailed results recorded in the NAPPA relational database. The software is fully integrated with the DNA Factory to provide tracking of each step in DNA purification and quantification (see workflow of the DNA purification step below). The slide production module provides a visual tool to help researchers rapidly design and build high-density microarrays and generate the robot programs needed to produce these microarray slides in a high-throughput environment. The experiment results module helps scientists to design effective NAPPA experiments and to track high-throughput results. The final module provides powerful data analysis and visualization tools to enable researchers to effectively glean information from the vast data repository to help ensure high quality experiments and accelerate research.
Other innovations
Our team is always innovating to enable scientific progress. These innovations include affinity reagents, NMR and protein purification.
- Affinity reagents. High quality protein capture reagents are critical for the elucidation of protein function, molecular diagnostics and even therapeutics. Recent acceleration in protein discovery will require affinity reagents to nearly all proteins. To achieve this goal, we have developed a new and innovative concept for creating bivalent affinity reagents called NuPromers.
Our pipeline begins with a high-throughput protein production system for generating target proteins followed by a high throughput selection for high affinity (typically low nanomolar) peptides using a state-of-the-art mRNA display system capable of searching libraries of >1012 different peptides.
Peptides produced by the selection are ranked by Surface Plasmon Resonance Imaging (SPRi) to identify candidate ligands with the highest affinity and specificity for their cognate protein. High quality peptides are then examined in pairwise combinations on synthetic DNA scaffolds using our ligand interactions by nucleotide conjugates (LINC) technology to identify the optimal peptide pair and linker (distance and geometry) required to produce a NuPromer. Our preliminary data demonstrates that NuPromers are straightforward to produce and bind their targets with low to sub-nanomolar affinity. Moreover, straightforward chemical approaches make NuPromers a cost-effective alternative to traditional antibodies that can be produced in gram quantities with readily available and quality-controlled methods. We believe our platform and bivalent affinity reagents will have a broad application in protein function, molecular diagnostics and therapeutics.
- Nuclear magnetic resonance. Membrane proteins, as a class, make up the majority of therapeutic targets and play essential roles in biology and pathophysiology. We investigate the structure, dynamics and function of this class of proteins with an interdisciplinary approach synthesizing the output from multiple disparate techniques which allows us to tackle challenging yet biomedically relevant problems. Our lab has interest in fundamental questions related to structural synthetic biology, especially in the following areas:
- TRP channel gating and modulation.
- Structural synthetic biology.
- Membrane protein structural enzymology.
- Methods development for
- Protein purification. We have designed a mammalian protein production pipeline for full-length proteins that utilizes recently developed in vitro protein expression systems. We typically employ multiple expression systems to achieve a high (>80%) success rate using E. coli, cell-free wheat germ extract (WGE) or cell-free human HeLa cell extract (HCE). We have adapted a HALO-tag based protein purification protocol followed by protease cleavage to ensure high purity. This pipeline has been automated with liquid handling robots, allowing for the purification of 96 proteins in parallel. Yields of purified proteins range from several to 10s of micrograms. We have successfully applied proteins produced from our system to the discovery and characterization of novel affinity reagents for the human proteome.
Core services
A powerful suite of services and bioinformatics tools are available, serving both our activities and the broader scientific community. Center researchers have been fortunate to work with many bioinformaticians and programmers, combining their varied talents in order to write programs and design databases that support our High Throughput Cloning Core, including programs that track all of the samples in the lab (FLEXGene), automatically analyze sequencing results (ACE), and store data and track our plasmid samples (DNASU). In addition, we have developed programs to track and help design NAPPA experiments (NAPPA Tracking Database) and to do comprehensive literature searches and analysis (MedGene and BioGene).
The core services in the CPD that are supported by these state-of-the-art informatics include the DNASU Plasmid Repository that distributes plasmids to researchers worldwide. Our production team is expert at cloning or transferring genes in high throughput for grant-funded projects in our center, which supports both our researchers and our plasmid repository. Finally, our NAPPA Protein Array Core offers high throughput DNA preparation, preset and custom protein array production, and array screening services.
These services further establish us as a central clearing house for advanced research into the human proteome.
- DNASU plasmids repository.
- DNASU sequencing.
- NAPPA protein array core.
- Bioinformatics.
- High throughput cloning.
Support the Biodesign Virginia G. Piper Center for Personalized Diagnostics
Developing new diagnostic tools to pinpoint the molecular manifestations of disease based on individual patient profiles.